My first tutorial: pythonPalIcoFoam
You can watch a short videotutorial for this case, here.
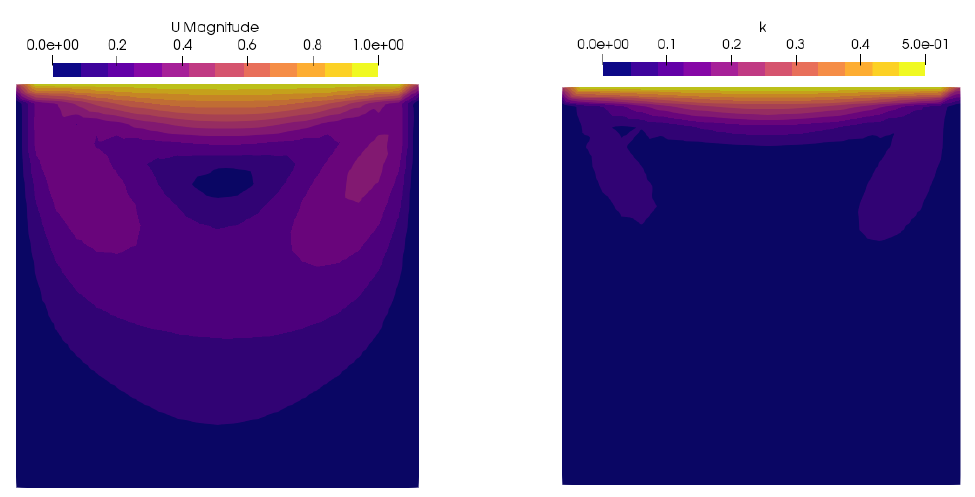
In this tutorial, we create a modified version of icoFoam called pythonPalIcoFoam, where we use pythonPal to calculate the specific kinetic energy, \(k = \frac {1}{2} U \cdot U\)
Provided that pybind11 is installed (see installing), the steps to go from icoFoam to pythonPalIcoFoam are:
-
Copy the
icoFoamdirectory to a directorypythonPalIcoFoam, then rename the .C file inside it and the C file and executable file names insideMake/files( Just replaceicoFoamwithpythonPalIcoFoam). -
Copy the
pythonPal.Hfile to thepythonPalIcoFoamdirectory. -
Include
pythonPal.Hat the top ofpythonPalIcoFoam.C;
#include "pythonPal.H"
- In
createFields.H, construct avolScalarFieldcalledk, initialised to zero;
volScalarField k
(
IOobject
(
"k",
mesh.time().timeName(),
mesh,
IOobject::READ_IF_PRESENT,
IOobject::AUTO_WRITE
),
mesh,
dimensionedScalar("k", dimensionSet(0, 2, -2, 0, 0, 0, 0), 0.0)
);
- In the parent folder, create a Python file and define a function that receives the
Ufield from OpenFOAM and calculatesk. We called that filepython_script.pyand the functioncalculatek.
def calculatek(field):
return np.sum(field * field, axis = 1)[:, np.newaxis] / 2
- In
pythonPalIcoFoam.C, create an object of typepythonPaland load the Python filepython_script.py:
pythonPal myPythonPal("python_script.py", true);
- At the end of the time-loop, pass the
Uandkfields to Python viapythonPal:
myPythonPal.passToPython(U, "U");
myPythonPal.passToPython(k, "k");
- Calculate
kviapythonPal:
myPythonPal.execute("k[:, :] = calculatek(U)");
- Finally, write the
kfield to disk, e.g. for viewing in ParaView.
k.write();
Figure shows the cavity case's velocity magnitude and specific kinetic energy field results.
To learn more about how pythonPal works, see How Does It Work?.